今天小果想对对3组及以上的亚组进行生存分析,并打印配对比较的统计表格,代码如下:
- 安装需要的R包
install.packages(“RcolorBrewer”)
install.packages(“tibble”)
install.packages(“ggpp”)
install.packages(“survival”)
install.packages(“survminer”)
- 导入需要的R包
library(survival)
library(survminer)
library(RColorBrewer)
library(tibble)
library(ggpp)
- 代码展示
#导入数据
dat<-read.table(“survival.txt”,sep=”\t”,row.names=1,check.names=F,stringsAsFactors = F,header = T)
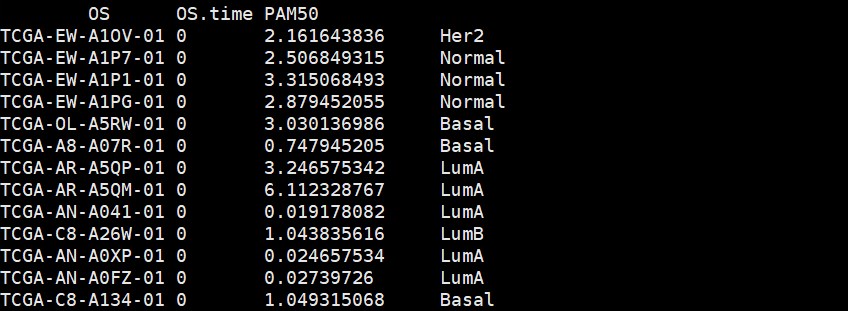
#将PAM50分型按照ABCDE标记
dat$group <- sapply(dat$PAM50,function(x) {
switch(x,
“Basal” = “A”, # 将Basal标记为A
“Her2” = “B”, # 将Her2标记为B
“LumA” = “C”, # 将LumA标记为C
“LumB” = “D”, # 将LumB标记为D
“Normal” = “E”)}) # 将Normal标记为E
##将生存时间转换为月份
dat$OS.time <- dat$OS.time * 12
# 生存分析
fitd <- survdiff(Surv(OS.time, OS) ~ group,
data = dat,
na.action = na.exclude)
p.val <- 1 – pchisq(fitd$chisq, length(fitd$n) – 1)
fit <- survfit(Surv(OS.time, OS)~ group,
data = dat,
type = “kaplan-meier”,
error = “greenwood”,
conf.type = “plain”,
na.action = na.exclude)
# 配对生存分析
ps <- pairwise_survdiff(Surv(OS.time, OS)~ group,
data = dat,
p.adjust.method = “none”) # 这里不使用矫正,若需要矫正可以将none替换为BH
# 设置颜色
mycol <- brewer.pal(n = 10, “Paired”)[c(2,4,6,8,10)]
# 绘制基础图形
## 隐藏类标记
names(fit$strata) <- gsub(“group=”, “”, names(fit$strata))
## 生存曲线图
p <- ggsurvplot(fit = fit,
conf.int = FALSE, # 不绘制置信区间
risk.table = TRUE, # 生存风险表
risk.table.col = “strata”,
palette = mycol, # KM曲线颜色
data = dat,
xlim = c(0,120), # 时间轴,一般考虑5年(原文)或者10年长度
size = 1,
break.time.by = 12, # 时间轴的刻度(每年)
legend.title = “”,
xlab = “Time (months)”,
ylab = “Overall survival”,
risk.table.y.text = FALSE,
tables.height = 0.3) # 风险表的高度
## 添加overall pvalue
p.lab <- paste0(“log-rank test P”,
ifelse(p.val < 0.001, ” < 0.001″, # 若P值<0.001则标记为“<0.001”
paste0(” = “,round(p.val, 3))))
p$plot <- p$plot + annotate(“text”,
x = 0, y = 0.55, # 在y=0.55处打印overall p值
hjust = 0,
fontface = 4,
label = p.lab)
## 添加配对表格
addTab <- as.data.frame(as.matrix(ifelse(round(ps$p.value, 3) < 0.001, “<0.001”,
round(ps$p.value, 3))))
addTab[is.na(addTab)] <- “-“
df <- tibble(x = 0, y = 0, tb = list(addTab))
p$plot <- p$plot +
geom_table(data = df,
aes(x = x, y = y, label = tb),
table.rownames = TRUE)
##生成图片
pdf(“km_curve_with_pairwise_logrank.pdf”, width = 4.5, height = 6)
print(p)
dev.off()
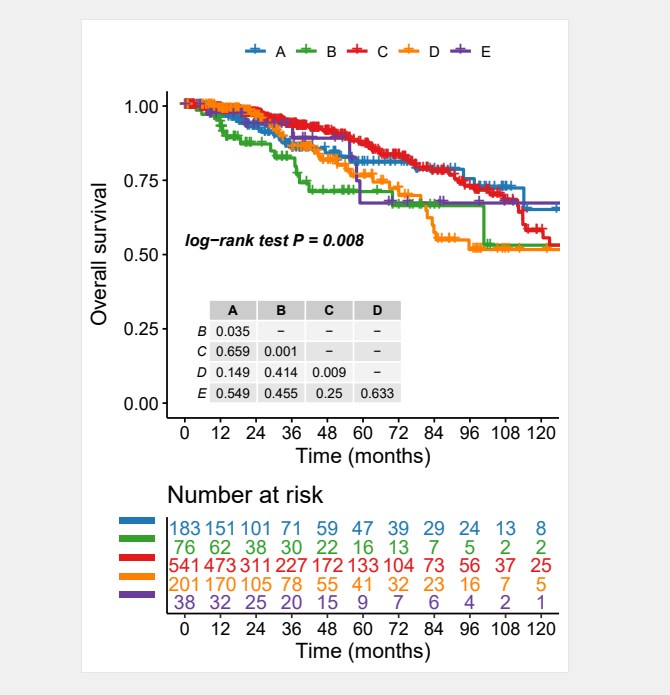
最终对多个分组的数据进行了分析,绘制了添加配对比较的文件的图片,看起来效果还不错。
对多个分组进行生存分析
今天小果想对对3组及以上的亚组进行生存分析,并打印配对比较的统计表格,代码如下:
- 安装需要的R包
install.packages(“RcolorBrewer”)
install.packages(“tibble”)
install.packages(“ggpp”)
install.packages(“survival”)
install.packages(“survminer”)
- 导入需要的R包
library(survival)
library(survminer)
library(RColorBrewer)
library(tibble)
library(ggpp)
- 代码展示
#导入数据
dat<-read.table(“survival.txt”,sep=”\t”,row.names=1,check.names=F,stringsAsFactors = F,header = T)

#将PAM50分型按照ABCDE标记
dat$group <- sapply(dat$PAM50,function(x) {
switch(x,
“Basal” = “A”, # 将Basal标记为A
“Her2” = “B”, # 将Her2标记为B
“LumA” = “C”, # 将LumA标记为C
“LumB” = “D”, # 将LumB标记为D
“Normal” = “E”)}) # 将Normal标记为E
##将生存时间转换为月份
dat$OS.time <- dat$OS.time * 12
# 生存分析
fitd <- survdiff(Surv(OS.time, OS) ~ group,
data = dat,
na.action = na.exclude)
p.val <- 1 – pchisq(fitd$chisq, length(fitd$n) – 1)
fit <- survfit(Surv(OS.time, OS)~ group,
data = dat,
type = “kaplan-meier”,
error = “greenwood”,
conf.type = “plain”,
na.action = na.exclude)
# 配对生存分析
ps <- pairwise_survdiff(Surv(OS.time, OS)~ group,
data = dat,
p.adjust.method = “none”) # 这里不使用矫正,若需要矫正可以将none替换为BH
# 设置颜色
mycol <- brewer.pal(n = 10, “Paired”)[c(2,4,6,8,10)]
# 绘制基础图形
## 隐藏类标记
names(fit$strata) <- gsub(“group=”, “”, names(fit$strata))
## 生存曲线图
p <- ggsurvplot(fit = fit,
conf.int = FALSE, # 不绘制置信区间
risk.table = TRUE, # 生存风险表
risk.table.col = “strata”,
palette = mycol, # KM曲线颜色
data = dat,
xlim = c(0,120), # 时间轴,一般考虑5年(原文)或者10年长度
size = 1,
break.time.by = 12, # 时间轴的刻度(每年)
legend.title = “”,
xlab = “Time (months)”,
ylab = “Overall survival”,
risk.table.y.text = FALSE,
tables.height = 0.3) # 风险表的高度
## 添加overall pvalue
p.lab <- paste0(“log-rank test P”,
ifelse(p.val < 0.001, ” < 0.001″, # 若P值<0.001则标记为“<0.001”
paste0(” = “,round(p.val, 3))))
p$plot <- p$plot + annotate(“text”,
x = 0, y = 0.55, # 在y=0.55处打印overall p值
hjust = 0,
fontface = 4,
label = p.lab)
## 添加配对表格
addTab <- as.data.frame(as.matrix(ifelse(round(ps$p.value, 3) < 0.001, “<0.001”,
round(ps$p.value, 3))))
addTab[is.na(addTab)] <- “-“
df <- tibble(x = 0, y = 0, tb = list(addTab))
p$plot <- p$plot +
geom_table(data = df,
aes(x = x, y = y, label = tb),
table.rownames = TRUE)
##生成图片
pdf(“km_curve_with_pairwise_logrank.pdf”, width = 4.5, height = 6)
print(p)
dev.off()

最终对多个分组的数据进行了分析,绘制了添加配对比较的文件的图片,看起来效果还不错。