Limma分析还能这么用?2步教你用limma分析找出最佳的靶向基因!


公众号后台回复“111”
领取本篇代码、基因集或示例数据等文件
文件编号:240115
需要租赁服务器的小伙伴可以扫码添加小果,此外小果还提供生信分析,思路设计,文献复现等,有需要的小伙伴欢迎来撩~

##### 导入R包 #####library(limma)library(splines)library(splines2)ns<-naturalSplinelibrary(scales)library(BinfTools)##### 构建功能函数 ########## 根据给定的细胞系,构建一个线性模型矩阵,用于后续的差异分析 #####makemodel<-function(x, colname, cell.lines){cell.lines<-sapply(strsplit(cell.lines, split="_", fixed=T),'[[', 1)if(ncol(x)==1){olCells<-rownames(x)[which(rownames(x) %in% cell.lines)]x<-as.data.frame(x[olCells,])rownames(x)<-olCellscolnames(x)<-colname} else {x<-x[which(rownames(x) %in% cell.lines),]}sm<-NULLif(ncol(x)==1){sm<-ns(x[,1])} else {sm<-ns(x[,which(colnames(x) %in% colname)])}dm<-model.matrix(~sm)return(dm)}##### 根据给定的细胞系和表达矩阵,返回符合条件的细胞系名称 #####olCells<-function(cell.lines, x){tmp<-sapply(strsplit(cell.lines, split="_", fixed=T),'[[',1)#return(tmp)return(cell.lines[which(tmp %in% rownames(x))])}##### 根据给定的两个GSEA结果,进行差异分析和可视化,比较不同物种或条件下的基因集富集情况 #####compGSEA<-function(x, y, title="Comparing GSEA results", xlab="Human", ylab="Mouse", p=1){x<-subset(x, padj <p)< span></p)<>y<-subset(y, padj <p)< span></p)<>x<-x[which(x$pathway %in% y$pathway),]y<-y[which(y$pathway %in% x$pathway),]x<-x[order(x$pathway),]y<-y[order(y$pathway),]if(!all.equal(x$pathway, y$pathway)){stop("Check again...")}dat<-data.frame(row.names=x$pathway,x=x$NES,y=y$NES)dat$Direction<-ifelse(dat$x*dat$y>0, "Positive", "Negative")dat$Trend<-ifelse(dat$x*dat$y>0,abs(dat$x-dat$y), abs(dat$x-dat$y)*-1)#print(head(dat))g<-ggpubr::ggscatter(dat, x="x", y="y", add="reg.line", conf.int=T, color="Trend",#palette=c(Negative="blue",Positive="red"),cor.coef=T, add.params=list(color="purple",fill="purple"),xlab=paste(xlab,"NES"), ylab=paste(ylab,"NES")) +ggpubr::gradient_color(c("blue","white","red"))print(g)return(dat)}#### 提取基因的符号名称 ####putSym<-function(res){res$hgnc_symbol<-sapply(strsplit(rownames(res), split=".", fixed=T),'[[',1)return(res)}#### 规范化 ####fixRows<-function(x, obtype=c("res","counts"), col=NULL){if(obtype[1]=="res"){x<-x[order(x$baseMean, decreasing=T),]}if(obtype[1]=="counts"){x<-x[order(rowMeans(x), decreasing=T),]}genes<-c()if(is.null(col)){genes<-sapply(strsplit(rownames(x), ".",T),'[[',1)} else {genes<-x[,which(colnames(x) %in% col)]}dups<-unique(genes[which(duplicated(genes))])to_remove <- c()for (i in 1:length(dups)) {ind <- which(genes %in% dups[i], arr.ind = T)to_remove <- c(to_remove, ind[-1])}x <- x[-to_remove, ]genes<-genes[-to_remove]rownames(x)<-genesreturn(x)}##### 导入数据 ######读取CCLE(Cancer Cell Line Encyclopedia)的基因表达数据和细胞系信息数据CCLE<-read.csv("/data/CCLE_expression.csv")#区分每个细胞系info<-read.csv("/data/sample_info.csv")#通过 DepMap ID 和 CCLE 名称匹配细胞系标识符CCLE<-merge(CCLE, info[,c(1,4)], by.x="X", by.y="DepMap_ID")rownames(CCLE)<-CCLE$CCLE_NameCCLE<-CCLE[,-(which(colnames(CCLE) %in% c("X","DepMap_ID","CCLE_Name")))]CCLE<-t(CCLE)#读入细胞系注释数据(识别 SCLC 细胞系——小细胞肺癌)x<-read.delim("/data/Cell_lines_annotations_20181226.txt")SCLC<-CCLE[,which(colnames(CCLE) %in% x[which(x$type_refined %in% "lung_small_cell"),]$CCLE_ID)]#识别SCLC的子类ATOH1<-c("CORL24_LUNG","HCC33_LUNG")POU2F3<-c("NCIH1048_LUNG", "CORL311_LUNG","NCIH211_LUNG","NCIH526_LUNG")YAP1<-c("SW1271_LUNG","NCIH1341_LUNG","DMS114_LUNG", "NCIH196_LUNG", "NCIH1339_LUNG", "SBC5_LUNG","NCIH2286_LUNG","NCIH841_LUNG")YAP1_2<-c("SW1271_LUNG","NCIH1341_LUNG","DMS114_LUNG", "NCIH1339_LUNG", "SBC5_LUNG","NCIH2286_LUNG","NCIH841_LUNG") #YAP1_2 removes outlier sample NCIH196_LUNGNEUROD1<-c("NCIH2066_LUNG","CORL279_LUNG","NCIH1694_LUNG","NCIH524_LUNG","DMS273_LUNG","NCIH446_LUNG","NCIH2171_LUNG","NCIH82_LUNG","SCLC21H_LUNG")ASCL1<-colnames(SCLC)ASCL1<-ASCL1[which(!ASCL1 %in% ATOH1)]ASCL1<-ASCL1[which(!ASCL1 %in% POU2F3)]ASCL1<-ASCL1[which(!ASCL1 %in% YAP1)]ASCL1<-ASCL1[which(!ASCL1 %in% NEUROD1)]typing<-data.frame(Cell.line=c(ATOH1,POU2F3,YAP1,NEUROD1,ASCL1),type=c(rep("ATOH1", length(ATOH1)), rep("POU2F3", length(POU2F3)),rep("YAP1", length(YAP1)), rep("NEUROD1", length(NEUROD1)),rep("ASCL1", length(ASCL1))))#加载 TAK243 药物反应数据x<-read.csv("/data/TAK_final_results.csv", row.names=1)#有些细胞系我们有 TAK 数据,但在数据集中没有表达数据,所以仅保留同时具有这两种数据的细胞系x<-as.data.frame(x[which(rownames(x) %in% sapply(strsplit(colnames(CCLE), split="_", fixed=T),'[[',1)),])CCLE<-CCLE[,which(sapply(strsplit(colnames(CCLE),"_",T),'[[',1) %in% rownames(x))]#将药物反应数据中细胞系的顺序与表达数据匹配以进行表达建模x<-x[match(sapply(strsplit(colnames(CCLE), split="_", T),'[[',1), rownames(x)),]x<-x[complete.cases(x),]#检查所有细胞系的顺序是否相同all.equal(sapply(strsplit(colnames(CCLE), "_", T),'[[',1), rownames(x))

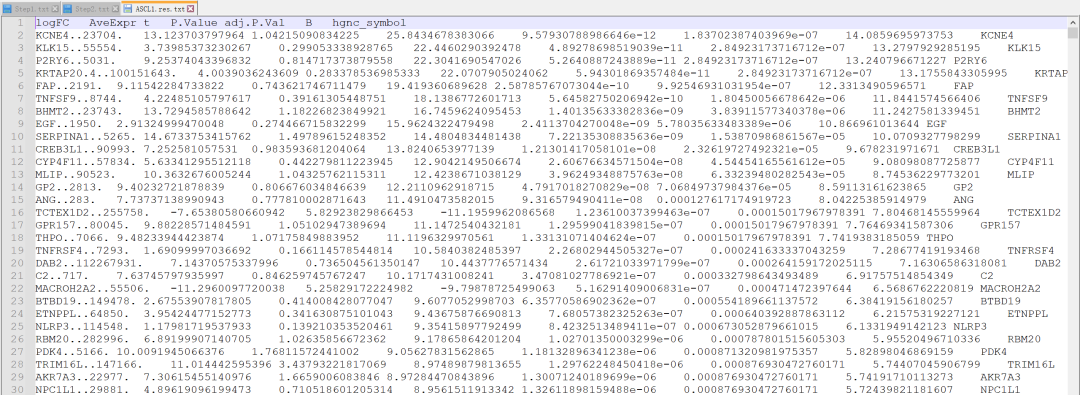
##### Limma 分析 ######为 EC50 值创建spline模型nsEC50<-ns(x$EC50)#为 Limma 制作设计模型矩阵dm_EC50<-model.matrix(~nsEC50) #Model for all cell lines#使用自定义函数为子类型组生成设计模型矩阵dm_EC50_YAP<-makemodel(x, "EC50", YAP1) #All YAP1-high/Triple Negativedm_EC50_YAP2<-makemodel(x, "EC50", YAP1_2) #All YAP1-high except NCIH196_LUNG (outlier)dm_EC50_ND1<-makemodel(x, "EC50", NEUROD1) #All NEUROD1-highdm_EC50_ASCL1<-makemodel(x, "EC50", ASCL1) #All ASCL1-high#表达矩阵子集仅包含每次分析所需的细胞系CCLE<-CCLE[,which(colnames(CCLE) %in% olCells(colnames(CCLE), x))] #All cell linesCCLE2<-CCLE[,which(!colnames(CCLE) %in% "NCIH196_LUNG")] #All cell lines except NCIH196_LUNG (outlier)dm_EC50_2<-makemodel(x[which(!rownames(x) %in% "NCIH196"),], "EC50", colnames(CCLE2)) #Create design model matrix for all SCLC cell lines except NCIH196_LUNG (outlier)CCLE_YAP<-CCLE[,which(colnames(CCLE) %in% olCells(YAP1, x))]CCLE_YAP2<-CCLE[,which(colnames(CCLE) %in% olCells(YAP1_2, x))]CCLE_ND1<-CCLE[,which(colnames(CCLE) %in% olCells(NEUROD1,x))]CCLE_ASCL1<-CCLE[,which(colnames(CCLE) %in% olCells(ASCL1,x))]#使用确定的 SCLC 亚型,使子集数据框仅包含已检测的 SCLC 细胞系subtyping<-subset(typing, Cell.line %in% colnames(CCLE))subtyping<-subtyping[match(colnames(CCLE), subtyping$Cell.line),] #Ensure same order of cell linesall.equal(subtyping$Cell.line, colnames(CCLE)) #Check - should return TRUE#创建条件向量subtype<-subtyping$typesubtype2<-subtype[-which(colnames(CCLE) %in% "NCIH196_LUNG")]#使用 Spearman 相关性检查样本如何关联和聚类#corHeat(CCLE, hmcol=colPal("Greys"),annodf=data.frame(row.names=subtyping$Cell.line, Subtype=subtyping$type), showTree=T)#运行 limma 分析 - 拟合线性回归模型EC50_fit<-lmFit(CCLE, design=dm_EC50)EC50_fit2<-lmFit(CCLE2, design=dm_EC50_2)EC50_fit_YAP<-lmFit(CCLE_YAP, design=dm_EC50_YAP)EC50_fit_YAP2<-lmFit(CCLE_YAP2, design=dm_EC50_YAP2)EC50_fit_ND1<-lmFit(CCLE_ND1, design=dm_EC50_ND1)EC50_fit_ASCL1<-lmFit(CCLE_ASCL1, design=dm_EC50_ASCL1)#差异表达的经验贝叶斯检验EC50_fit<-eBayes(EC50_fit)EC50_fit2<-eBayes(EC50_fit2)EC50_fit_YAP<-eBayes(EC50_fit_YAP)EC50_fit_YAP2<-eBayes(EC50_fit_YAP2)EC50_fit_ND1<-eBayes(EC50_fit_ND1)EC50_fit_ASCL1<-eBayes(EC50_fit_ASCL1)#返回所有结果(所有基因的回归系数和 p 值)EC50_res<-topTable(EC50_fit, number=Inf)EC50_res2<-topTable(EC50_fit2, number=Inf)EC50_res_YAP<-topTable(EC50_fit_YAP, number=Inf)EC50_res_YAP2<-topTable(EC50_fit_YAP2, number=Inf)EC50_res_ND1<-topTable(EC50_fit_ND1, number=Inf)EC50_res_ASCL1<-topTable(EC50_fit_ASCL1, number=Inf)#现在将包含基因符号的列 (hgnc_symbol) 添加到结果中EC50_res<-putSym(EC50_res)EC50_res2<-putSym(EC50_res2)EC50_res_ASCL1<-putSym(EC50_res_ASCL1)EC50_res_YAP<-putSym(EC50_res_YAP)EC50_res_YAP2<-putSym(EC50_res_YAP2)EC50_res_ND1<-putSym(EC50_res_ND1)#输出DE结果limResList<-list(SCLC_w196=EC50_res,SCLC_wo196=EC50_res2,YAP1_w196=EC50_res_YAP,YAP1_wo196=EC50_res_YAP2,ASCL1=EC50_res_ASCL1,NEUROD1=EC50_res_ND1)dir.create("/results/Limma_Results")mapply(write.table, limResList, file=paste0("/results/Limma_Results/",names(limResList),".res.txt"),quote=F, sep="t")
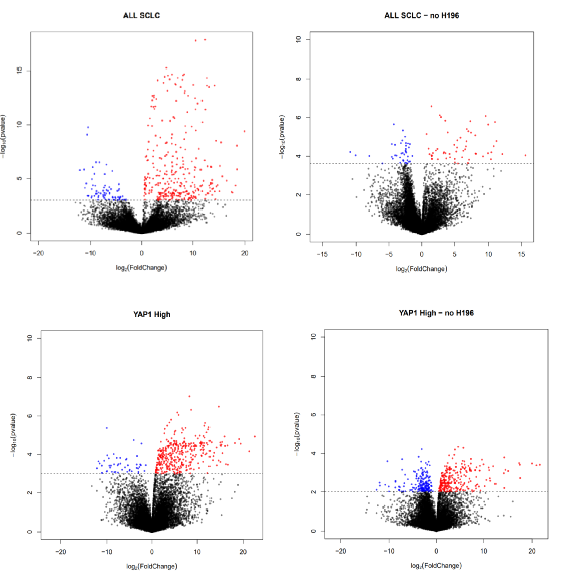
#将 limma 结果表转换为与 BinfTools 包函数兼容的格式#'fixRows()' will set the rownames to just the HGNC symbol instead of the HGNC.ENTREZID format it currently isbt_EC50<-fixRows(fromLimma(EC50_res), "res")bt_EC502<-fixRows(fromLimma(EC50_res2), "res")bt_EC50_YAP<-fixRows(fromLimma(EC50_res_YAP), "res")bt_EC50_YAP2<-fixRows(fromLimma(EC50_res_YAP2), "res")bt_EC50_ASCL1<-fixRows(fromLimma(EC50_res_ASCL1),"res")bt_EC50_ND1<-fixRows(fromLimma(EC50_res_ND1),"res")#用火山图显示显着的敏感/抵抗基因 (FDR < 0.05)#Save sensitizer(down/negatively correlated)/resistor(up/positively correlated) genespdf("/results/VolcanoPlots.pdf")EC_SCLC<-volcanoPlot(bt_EC50, "ALL SCLC", p=0.05, FC=0, showNum=F, returnDEG=T)EC_SCLC2<-volcanoPlot(bt_EC502, "ALL SCLC - no H196", p=0.05, FC=0, showNum=F, returnDEG=T)YAP_EC50<-volcanoPlot(bt_EC50_YAP, "YAP1 High", p=0.05, FC=0, showNum=F, returnDEG=T)YAP_EC502<-volcanoPlot(bt_EC50_YAP2, "YAP1 High - no H196", pval=0.01, FC=0, showNum=F, returnDEG=T)ASCL_EC50<-volcanoPlot(bt_EC50_ASCL1, "ASCL1 High", p=0.05, FC=0, showNum=F, returnDEG=T)ND1_EC50<-volcanoPlot(bt_EC50_ND1, "NEUROD1 High", pval=0.01, FC=0, showNum=F, returnDEG=T)dev.off()
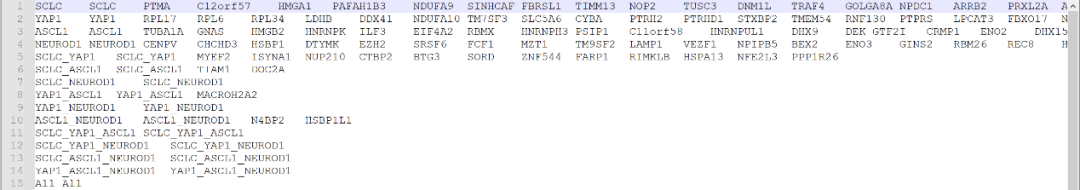
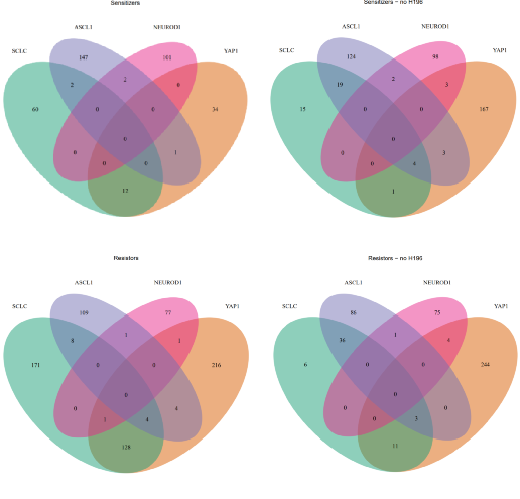
#输出维恩图并返回重叠基因pdf("/results/VennDiagrams.pdf")Sensitizers<-plotVenn(list(SCLC=EC_SCLC$Down, YAP1=YAP_EC50$Down,ASCL1=ASCL_EC50$Down, NEUROD1=ND1_EC50$Down),title="Sensitizers", retVals=T)Sensitizers2<-plotVenn(list(SCLC=EC_SCLC2$Down, YAP1=YAP_EC502$Down,ASCL1=ASCL_EC50$Down, NEUROD1=ND1_EC50$Down),title="Sensitizers - no H196", retVals=T)Resistors<-plotVenn(list(SCLC=EC_SCLC$Up, YAP1=YAP_EC50$Up,ASCL1=ASCL_EC50$Up, NEUROD1=ND1_EC50$Up),title="Resistors", retVals=T)Resistors2<-plotVenn(list(SCLC=EC_SCLC2$Up, YAP1=YAP_EC502$Up,ASCL1=ASCL_EC50$Up, NEUROD1=ND1_EC50$Up),title="Resistors - no H196", retVals=T)dev.off()#重命名重叠部分,使其更清晰OLnames<-c("SCLC","YAP1","ASCL1","NEUROD1","SCLC_YAP1", "SCLC_ASCL1","SCLC_NEUROD1","YAP1_ASCL1","YAP1_NEUROD1","ASCL1_NEUROD1","SCLC_YAP1_ASCL1","SCLC_YAP1_NEUROD1","SCLC_ASCL1_NEUROD1","YAP1_ASCL1_NEUROD1","All")names(Sensitizers)<-OLnamesnames(Resistors)<-OLnamesnames(Sensitizers2)<-OLnamesnames(Resistors2)<-OLnames#将重叠写出为基因集文件dir.create("/results/gmts/")write.gmt(Sensitizers, "/results/gmts/Sensitizer_Genes1.gmt")write.gmt(Sensitizers2, "/results/gmts/Sensitizer_Genes2.gmt")write.gmt(Resistors, "/results/gmts/Resistor_Genes1.gmt")write.gmt(Resistors2, "/results/gmts/Resistor_Genes2.gmt")
小果还提供思路设计、定制生信分析、文献思路复现;有需要的小伙伴欢迎直接扫码咨询小果,竭诚为您的科研助力!

定制生信分析
服务器租赁
扫码咨询小果


往期回顾
|
01 |
|
02 |
|
03 |
|
04 |