高分生信文章必备分析内容-Nomogram模型的构建和验证
今天小果为大家带来的分享内容为Nomogram模型的构建和验证,该分析在肿瘤生信文章中出现的频率非常高,小果本推文中主要绘制了列线图和校正曲线绘制,干货满满,非常值得小伙伴学习,接下来跟着小果马上开始今天的学习!
1.何为nomogram分析?
在进行分析之前,小果先为小伙伴介绍一下nomogram原理,nomogram实质就是回归方程的可视化,根据所有自变量回归系数的大小来制定评分标准,给每个自变量的每种取值水平一个评分,对每个样本就可计算得到一个总分,再通过得分与结局发生概率之间的转换函数来计算每个样本的结局时间发生的概率,我们通过Nomogram的方法得到的概率评价预测目标独立生存预后因素与预后间的相关性及对生存预后的预测关系。以上就是小果对nomogram的介绍,小伙伴们有没有理解呀!马上和小果开启实操练习吧!
公众号后台回复“111″,领取代码,代码编号:231005
2.准备需要的R包
#安装需要的R包install.packages("rms")install.packages("survival")#加载需要的R包library(rms)library(survival)

3.读取输入数据
#行名为样本名,列名为临床因素
pbc<-read.table("Nomogram.txt")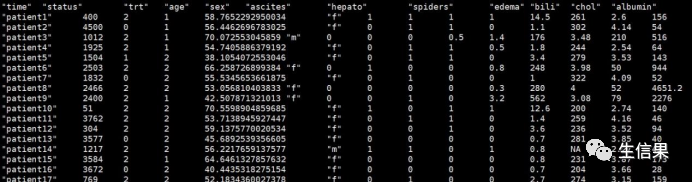
#对bili列进行数据分割pbc$catbili <- cut(pbc$bili,breaks=c(-Inf, 2, 4, Inf),labels=c("low","medium","high"))#增加新的列pbc$died <- pbc$status==24.Nomogram分析dd<-datadist(pbc)options(datadist="dd")options(na.action="na.delete")#多因素cox回归coxpbc<-cph(formula = Surv(time,died) ~ age + catbili + sex + copper + stage + trt ,data=pbc,x=T,y=T,surv = T,na.action=na.delete)surv<-Survival(coxpbc)#5年生存率surv3<-function(x) surv(1825,x)#8年生存率surv4<-function(x) surv(2920,x)x<-nomogram(coxpbc,fun = list(surv3,surv4),lp=T,funlabel = c('5-year survival Probability','8-year survival Probability'),maxscale = 100,fun.at = c(0.95,0.9,0.8,0.7,0.6,0.5,0.4,0.3,0.2,0.1))#绘制列线图pdf("nomogram_classical.pdf",width = 12,height = 10)plot(x, lplabel="Linear Predictor",xfrac=.35,varname.label=TRUE, varname.label.sep="=", ia.space=.2,tck=NA, tcl=-0.20, lmgp=0.3,points.label='Points', total.points.label='Total Points',total.sep.page=FALSE,cap.labels=FALSE,cex.var = 1.6,cex.axis = 1.05,lwd=5,label.every = 1,col.grid = gray(c(0.8, 0.95)))dev.off()
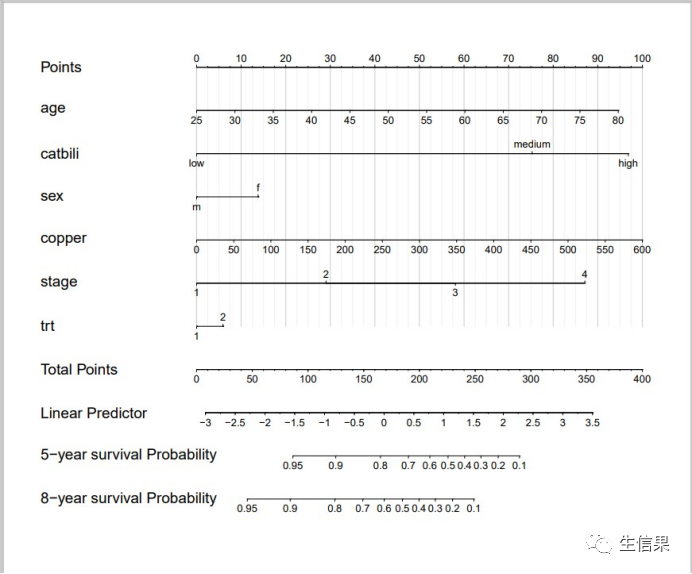
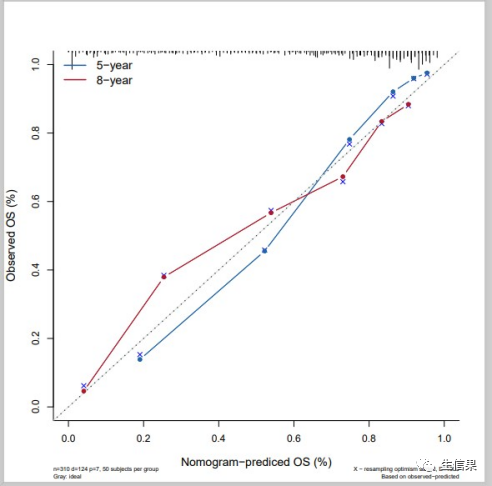
4.绘制校正曲线进行验证
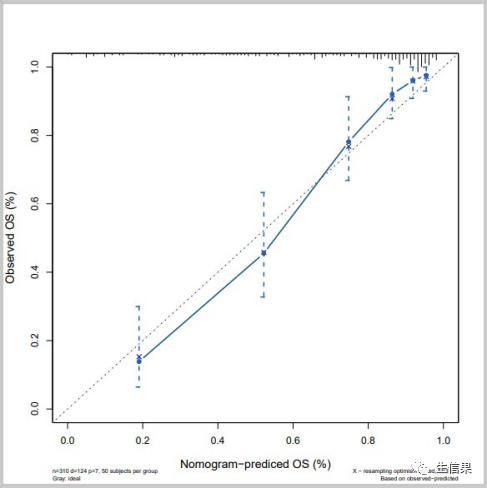
f5<-cph(formula = Surv(time,died) ~ age + catbili + sex + copper +stage + trt,data=pbc,x=T,y=T,surv = T,na.action=na.delete,time.inc = 1825)cal5<-calibrate(f5, cmethod="KM", method="boot",u=1825,m=50,B=1000)pdf("calibration_5y.pdf",width = 8,height = 8)plot(cal5,lwd = 2,lty = 2,errbar.col = c("#2166AC"),xlim = c(0,1),ylim= c(0,1),xlab = "Nomogram-prediced OS (%)",ylab = "Observed OS (%)",cex.lab=1.2, cex.axis=1, cex.main=1.2, cex.sub=0.6)lines(cal5[,c('mean.predicted',"KM")],type = 'b',lwd = 2,pch = 16,col = c("#2166AC"))mtext("")box(lwd = 1)abline(0,1,lty = 3,lwd = 2,col = c("#224444"))dev.off()
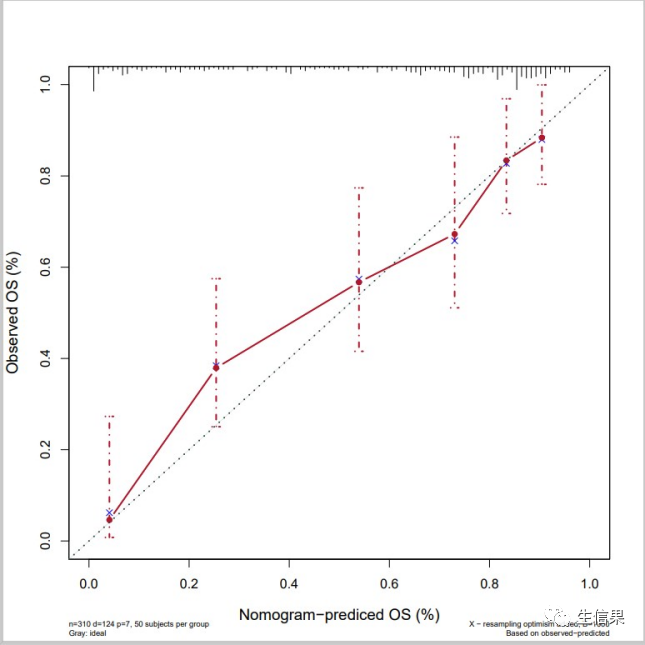
#8年生存率校正曲线,time.inc = 2920f8<-cph(formula = Surv(time,died) ~ age + catbili + sex + copper +stage + trt,data=pbc,x=T,y=T,surv = T,na.action=na.delete,time.inc = 2920)cal8<-calibrate(f8, cmethod="KM", method="boot",u=2920,m=50,B=1000)pdf("calibration_8y.pdf",width = 8,height = 8)plot(cal8,lwd = 2,lty = 4,errbar.col = c("#B2182B"),xlim = c(0,1),ylim= c(0,1),xlab = "Nomogram-prediced OS (%)",ylab = "Observed OS (%)",col = c("#B2182B"),cex.lab=1.2,cex.axis=1, cex.main=1.2, cex.sub=0.6)lines(cal8[,c('mean.predicted',"KM")],type= 'b',lwd = 2,col = c("#B2182B"),pch = 16)mtext("")box(lwd = 1)abline(0,1,lty= 3,lwd = 2,col =c("#224444"))dev.off()
#绘制5年和8年生存率校正曲线pdf("calibration_compare.pdf",width = 8,height = 8)plot(cal5,lwd = 2,lty = 0,errbar.col = c("#2166AC"),bty = "l", #只画左边和下边框xlim = c(0,1),ylim= c(0,1),xlab = "Nomogram-prediced OS (%)",ylab = "Observed OS (%)",col = c("#2166AC"),cex.lab=1.2,cex.axis=1, cex.main=1.2, cex.sub=0.6)lines(cal5[,c('mean.predicted',"KM")],type = 'b', lwd = 1, col = c("#2166AC"), pch = 16)mtext("")plot(cal8,lwd = 2,lty = 0,errbar.col = c("#B2182B"),xlim = c(0,1),ylim= c(0,1),col = c("#B2182B"),add = T)lines(cal8[,c('mean.predicted',"KM")],type = 'b', lwd = 1, col = c("#B2182B"), pch = 16)abline(0,1, lwd = 2, lty = 3, col = c("#224444"))legend("topleft", #图例的位置legend = c("5-year","8-year"), #图例文字col =c("#2166AC","#B2182B"), #图例线的颜色,与文字对应lwd = 2,#图例中线的粗细cex = 1.2,#图例字体大小bty = "n")#不显示图例边框dev.off()
今天小果完成了Nomogram模型的构建和验证,欢迎大家有问题与小果一起讨论哦
往期推荐

